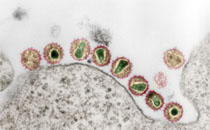
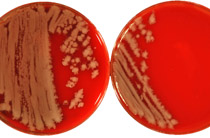
AG Retrovirus-Wirtszell-Interaktion
Leitung der AG: Prof. Dr. Norbert Bannert
Die Forschungsarbeiten befassen sich mit der Analyse retroviraler Replikationsmechanismen zur Aufklärung von Pathogenesemechanismen und der Entwicklung von Strategien zur Infektionsprävention. Einen weiteren Schwerpunkt bilden Untersuchungen zu zellulären Restriktionen der Proliferation endogener und exogener Retroviren mit Fokus auf der angeborenen Immunität.
In diesem Kontext wird aktuell an HIV-1 im Rahmen des ProNANOS-Projektes gearbeitet, für das Mittel aus dem zentralen Innovationsprogramm Mittelstand (ZIM) des Bundesministeriums für Wirtschaft und Klimaschutz eigeworben wurden. In dem Drittmittelprojekt wird die Rolle von Tetraspaninen während des Infektionsprozesses von HIV-1 analysiert. Die gewonnenen Erkenntnisse fließen in die Entwicklung einer innovativen tetraspaninbasierten Display-Technologie (tAnchor) zur Infektiosprävention und Diagnostik ein. In einem Promotionsprojektes wird darüber hinaus an der Identifizierung von Virulenzfaktoren von HIV-1 geforscht. Die Arbeiten zu endogenen Retroviren finden primär an HERV-K(HML-2) statt. HERV-K(HML-2) ist die jüngste und aktivste Familie humaner endogener Retroviren. Proteine und Viruspartikel sind in Tumorzellen zu finden und werden mit Prozessen der Krebsentwicklung, der Amyotrophen Lateralsklerose sowie diversen Autoimmunerkrankungen in Verbindung gebracht.
Im Zuge der Mitarbeit im DFG-Schwerpunktprogramm "Innate Sensing and Restriction of Retroviruses (SPP1923)" fokussieren wir auf interferoninduzierte zelluläre Sensoren und Restriktionsfaktoren sowie auf bestimmte antivirale Mechanismen gegen endogene und exogene Retroviren. Ein aktueller Schwerpunkt ist die Analyse der Restriktion von HERV-K(HML-2) durch IFI16 und andere Proteine der PYHIN-Familie. Ziel ist es, die Erkennung retroviraler Nukleinsäuren und Proteine durch zelluläre Infektionssensoren und der daraus folgenden Konsequenzen für die Transmission und Virusreplikation zu verstehen sowie gruppen- und subtypspezifische Unterschiede herauszuarbeiten.
In einem weiteren, aus Sonderforschungsmitteln des RKI finanzierten Projekt, werden gemeinsam mit Mitarbeitern des Fachgebiets 34 Immunantworten gegen SARS-CoV-2-Infektionen und -Vakzinierungen bei HIV-Patienten untersucht.
Die Arbeiten des Teams sollen zu einem tieferen Verständnis der komplexen Virus-Wirt-Interaktionen führen. Die gewonnenen Erkenntnisse werden dazu beitragen, die Public Health-Relevanz der Retroviren genauer einschätzen zu können sowie potenzielle Angriffspunkte für neue Präventionsmaßnahmen und therapeutische Interventionen aufzeigen.
Anfragen zu Praktika sowie Master- und Bachelorarbeiten bitte an die Arbeitsgruppenleitung richten.
Publikationen von Mitarbeitern der Arbeitsgruppe (2013-2024, Peer-Review)
- Ivanusic D, Maier J, Icli S, Falcone V, Bernauer H, Bannert N: tANCHOR-cell-based assay for monitoring of SARS-CoV-2 neutralizing antibodies rapidly adaptive to various receptor-binding domains. iScience. 2024 Feb 5;27(3):109123.
- Bernauer H, Maier J, Bannert N, Ivanusic D: tANCHOR cell-based ELISA approach as a surrogate for antigen-coated plates to monitor specific IgG directed to the SARS-CoV-2 receptor-binding domain. Biol Methods Protoc. 2024 Jan 19;9(1).
- Köppke J, Keller LE, Stuck M, Arnow ND, Bannert N, Doellinger J, Cingöz O: Direct translation of incoming retroviral genomes. Nat Commun. 2024 Jan 5;15(1):299.
- Toudic C, Maurer M, St-Pierre G, Xiao Y, Bannert N, Lafond J, Rassart É, Sato S, Barbeau B: Galectin-1 Modulates the Fusogenic Activity of Placental Endogenous Retroviral Envelopes. Viruses. 2023 Dec 16;15(12):2441.
- Bernauer H, Schlör A, Maier J, Bannert N, Hanack K, Ivanusic D: tANCHOR fast and cost-effective cell-based immunization approach with focus on the receptor-binding domain of SARS-CoV-2. Biol Methods Protoc. 2023 Dec 12;8(1).
- Fiebig U, Altmann B, Hauser A, Koppe U, Hanke K, Gunsenheimer-Bartmeyer B, Bremer V, Baumgarten A, Bannert N: Transmitted drug resistance and subtype patterns of viruses from reported new HIV diagnoses in Germany, 2017-2020. BMC Infect Dis. 2023 Oct 10;23(1):673.
- Krings A, Schmidt D, Kollan C, Meixenberger K, Bannert N, Münstermann D, Tiemann C, Bremer V, Gunsenheimer-Bartmeyer B: Increasing hepatitis B vaccination coverage and decreasing hepatitis B co-infection prevalence among people with HIV-1 in Germany, 1996-2019. Results from a cohort study primarily in men who have sex with men. German HIV-1 Seroconverter Study Group. HIV Med. 2024 Feb;25(2):201-211.
- Fiebig U, Altmann B, Hauser A, Koppe U, Hanke K, Gunsenheimer-Bartmeyer B, Bremer V, Baumgarten A, Bannert N: Primary Resistance of Viruses in Reported New Cases of HIV, 2017-2020. Dtsch Arztebl Int. 2023 Jul 10;120(27-28):479-480.
- Zhao L, Wymant C, Blanquart F, Golubchik T, Gall A, Bakker M, Bezemer D, Hall M, Ong SH, Albert J et al: Phylogenetic estimation of the viral fitness landscape of HIV-1 set-point viral load. Virus Evol 2022, 8(1):veac022.
- Wymant C, Bezemer D, Blanquart F, Ferretti L, Gall A, Hall M, Golubchik T, Bakker M, Ong SH, Zhao L, .., Bannert N, et al: A highly virulent variant of HIV-1 circulating in the Netherlands. Science 2022, 375(6580):540-545.
- Pantke A, Hoebel J, An der Heiden M, Michalski N, Gunsenheimer-Bartmeyer B, Hanke K, Bannert N, Bremer V, Koppe U: The impact of regional socioeconomic deprivation on the timing of HIV diagnosis: a cross-sectional study in Germany. BMC Infect Dis 2022, 22(1):258.
- Krings A, Schmidt D, Meixenberger K, Bannert N, Munstermann D, Tiemann C, Kollan C, Gunsenheimer-Bartmeyer B: Decreasing prevalence and stagnating incidence of Hepatitis C-co-infection among a cohort of HIV-1-positive patients, with a majority of men who have sex with men, in Germany, 1996-2019. J Viral Hepat 2022, 29(6):465-473.
- Ivanusic D, Madela K, Bannert N, Denner J: Time-lapse imaging of CD63 dynamics at the HIV-1 virological synapse by using agar pads. MicroPubl Biol 2022, 2022.
- Al-Shehabi H, Bannert N: PERV induces CXCL10 in human monocytes and monocyte-derived primary cells. Intervirology 2022.
- Ivanusic D, Madela K, Burghard H, Holland G, Laue M, Bannert N: tANCHOR: a novel mammalian cell surface peptide display system. Biotechniques 2021, 70(1):21-28.
- Ivanusic D, Madela K, Bannert N, Denner J: The large extracellular loop of CD63 interacts with gp41 of HIV-1 and is essential for establishing the virological synapse. Sci Rep 2021, 11(1):10011.
- Cingoz O, Arnow ND, Puig Torrents M, Bannert N: Vpx enhances innate immune responses independently of SAMHD1 during HIV-1 infection. Retrovirology 2021, 18(1):4.
- Wratil PR, Rabenau HF, Eberle J, Stern M, Munchhoff M, Friedrichs I, Sturmer M, Berger A, Kuttner-May S, Munstermann D et al: Comparative multi-assay evaluation of Determine HIV-1/2 Ag/Ab Combo rapid diagnostic tests in acute and chronic HIV infection. Med Microbiol Immunol 2020, 209(2):139-150.
- Hanke K, Fiedler S, Grumann C, Ratmann O, Hauser A, Klink P, Meixenberger K, Altmann B, Zimmermann R, Marcus U et al: A Recent Human Immunodeficiency Virus Outbreak Among People Who Inject Drugs in Munich, Germany, Is Associated With Consumption of Synthetic Cathinones. Open Forum Infect Dis 2020, 7(6):ofaa192.
- Eshetu A, Hauser A, Schmidt D, Bartmeyer B, Bremer V, Obermeier M, Ehret R, Volkwein A, Bock CT, Bannert N: Comparison of two immunoassays for concurrent detection of HCV antigen and antibodies among HIV/HCV co-infected patients in dried serum/plasma spots. J Virol Methods 2020, 279:113839.
- Eshetu A, Hauser A, An der Heiden M, Schmidt D, Meixenberger K, Ross S, Obermeier M, Ehret R, Bock CT, Bartmeyer B et al: Establishment of an anti-hepatitis C virus IgG avidity test for dried serum/plasma spots. J Immunol Methods 2020, 479:112744.
- Desimmie BA, Weydert C, Schrijvers R, Vets S, Demeulemeester J, Proost P, Paron I, De Rijck J, Mast J, Bannert N et al: Correction to: HIV-1 IN/Pol recruits LEDGF/p75 into viral particles. Retrovirology 2020, 17(1):23.
- Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J et al: Correction to: LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 2020, 17(1):22.
- Wang B, Kruger L, Machnowska P, Eshetu A, Gunsenheimer-Bartmeyer B, Bremer V, Hauser A, Bannert N, Bock CT: Characterization of a hepatitis C virus genotype 1 divergent isolate from an HIV-1 coinfected individual in Germany assigned to a new subtype 1o. Virol J 2019, 16(1):28.
- van de Laar MJ, Bosman A, Pharris A, Andersson E, Assoumou L, Ay E, Bannert N, Bartmeyer B, Brady M, Chaix ML et al: Piloting a surveillance system for HIV drug resistance in the European Union. Euro Surveill 2019, 24(19).
- Toudic C, Vargas A, Xiao Y, St-Pierre G, Bannert N, Lafond J, Rassart E, Sato S, Barbeau B: Galectin-1 interacts with the human endogenous retroviral envelope protein syncytin-2 and potentiates trophoblast fusion in humans. FASEB J 2019, 33(11):12873-12887.
- Paraskevis D, Beloukas A, Stasinos K, Pantazis N, de Mendoza C, Bannert N, Meyer L, Zangerle R, Gill J, Prins M et al: HIV-1 molecular transmission clusters in nine European countries and Canada: association with demographic and clinical factors. BMC Med 2019, 17(1):4.
- Machnowska P, Meixenberger K, Schmidt D, Jessen H, Hillenbrand H, Gunsenheimer-Bartmeyer B, Hamouda O, Kucherer C, Bannert N, German HIVSSG: Prevalence and persistence of transmitted drug resistance mutations in the German HIV-1 Seroconverter Study Cohort. PLoS One 2019, 14(1):e0209605.
- Hauser A, Heiden MA, Meixenberger K, Han O, Fiedler S, Hanke K, Koppe U, Hofmann A, Bremer V, Bartmeyer B et al: Evaluation of a BioRad Avidity assay for identification of recent HIV-1 infections using dried serum or plasma spots. J Virol Methods 2019, 266:114-120.
- Hanke K, Faria NR, Kuhnert D, Yousef KP, Hauser A, Meixenberger K, Hofmann A, Bremer V, Bartmeyer B, Pybus O et al: Reconstruction of the Genetic History and the Current Spread of HIV-1 Subtype A in Germany. J Virol 2019, 93(12).
- Gassowski M, Nielsen S, Bannert N, Bock CT, Bremer V, Ross RS, Wenz B, Marcus U, Zimmermann R, Group DS: History of detention and the risk of hepatitis C among people who inject drugs in Germany. Int J Infect Dis 2019, 81:100-106.
- Al-Shehabi H, Fiebig U, Kutzner J, Denner J, Schaller T, Bannert N, Hofmann H: Human SAMHD1 restricts the xenotransplantation relevant porcine endogenous retrovirus (PERV) in non-dividing cells. J Gen Virol 2019, 100(4):656-661.
- Wymant C, Blanquart F, Golubchik T, Gall A, Bakker M, Bezemer D, Croucher NJ, Hall M, Hillebregt M, Ong SH et al: Easy and accurate reconstruction of whole HIV genomes from short-read sequence data with shiver. Virus Evol 2018, 4(1):vey007.
- Olson A, Bannert N, Sonnerborg A, de Mendoza C, Price M, Zangerle R, Chaix ML, Prins M, Kran AB, Gill J et al: Temporal trends of transmitted HIV drug resistance in a multinational seroconversion cohort. AIDS 2018, 32(2):161-169.
- Ivanusic D, Pietsch H, Konig J, Denner J: Absence of IL-10 production by human PBMCs co-cultivated with human cells expressing or secreting retroviral immunosuppressive domains. PLoS One 2018, 13(7):e0200570.
- Haussig JM, Nielsen S, Gassowski M, Bremer V, Marcus U, Wenz B, Bannert N, Bock CT, Zimmermann R, group Ds: A large proportion of people who inject drugs are susceptible to hepatitis B: Results from a bio-behavioural study in eight German cities. Int J Infect Dis 2018, 66:5-13.
- Hauser A, Meixenberger K, Machnowska P, Fiedler S, Hanke K, Hofmann A, Bartmeyer B, Bremer V, Bannert N, Kuecherer C: Robust and sensitive subtype-generic HIV-1 pol genotyping for use with dried serum spots in epidemiological studies. J Virol Methods 2018, 259:32-38.
- Hauser A, Hofmann A, Meixenberger K, Altmann B, Hanke K, Bremer V, Bartmeyer B, Bannert N: Increasing proportions of HIV-1 non-B subtypes and of NNRTI resistance between 2013 and 2016 in Germany: Results from the national molecular surveillance of new HIV-diagnoses. PLoS One 2018, 13(11):e0206234.
- Derks L, Gassowski M, Nielsen S, An der Heiden M, Bannert N, Bock CT, Bremer V, Kucherer C, Ross S, Wenz B et al: Risk behaviours and viral infections among drug injecting migrants from the former Soviet Union in Germany: Results from the DRUCK-study. Int J Drug Policy 2018, 59:54-62.
- Bannert N, Hofmann H, Block A, Hohn O: HERVs New Role in Cancer: From Accused Perpetrators to Cheerful Protectors. Front Microbiol 2018, 9:178.
- Meixenberger K, Yousef KP, Smith MR, Somogyi S, Fiedler S, Bartmeyer B, Hamouda O, Bannert N, von Kleist M, Kucherer C: Molecular evolution of HIV-1 integrase during the 20 years prior to the first approval of integrase inhibitors. Virol J 2017, 14(1):223.
- McLaren PJ, Pulit SL, Gurdasani D, Bartha I, Shea PR, Pomilla C, Gupta N, Gkrania-Klotsas E, Young EH, Bannert N et al: Evaluating the Impact of Functional Genetic Variation on HIV-1 Control. J Infect Dis 2017, 216(9):1063-1069.
- Machnowska P, Hauser A, Meixenberger K, Altmann B, Bannert N, Rempis E, Schnack A, Decker S, Braun V, Busingye P et al: Decreased emergence of HIV-1 drug resistance mutations in a cohort of Ugandan women initiating option B+ for PMTCT. PLoS One 2017, 12(5):e0178297.
- Ivanusic D, Madela K, Denner J: Easy and low-cost stable positioning of suspension cells during live-cell imaging. J Biol Methods 2017, 4(4):e80.
- Hohn O, Norley S, Kucherer C, Bazarbachi A, El Hajj H, Marcus U, Zimmermann R, Bannert N: No significant HTLV seroprevalence in German people who inject drugs. PLoS One 2017, 12(8):e0183496.
- Hofmann A, Hauser A, Zimmermann R, Santos-Hovener C, Batzing-Feigenbaum J, Wildner S, Kucherer C, Bannert N, Hamouda O, Bremer V et al: Surveillance of recent HIV infections among newly diagnosed HIV cases in Germany between 2008 and 2014. BMC Infect Dis 2017, 17(1):484.
- Hauser A, Hofmann A, Hanke K, Bremer V, Bartmeyer B, Kuecherer C, Bannert N: National molecular surveillance of recently acquired HIV infections in Germany, 2013 to 2014. Euro Surveill 2017, 22(2).
- Fiebig U, Holzer A, Ivanusic D, Plotzki E, Hengel H, Neipel F, Denner J: Antibody Cross-Reactivity between Porcine Cytomegalovirus (PCMV) and Human Herpesvirus-6 (HHV-6). Viruses 2017, 9(11).
- Blanquart F, Wymant C, Cornelissen M, Gall A, Bakker M, Bezemer D, Hall M, Hillebregt M, Ong SH, Albert J et al: Correction: Viral genetic variation accounts for a third of variability in HIV-1 set-point viral load in Europe. PLoS Biol 2017, 15(7):e1002608.
- Blanquart F, Wymant C, Cornelissen M, Gall A, Bakker M, Bezemer D, Hall M, Hillebregt M, Ong SH, Albert J et al: Viral genetic variation accounts for a third of variability in HIV-1 set-point viral load in Europe. PLoS Biol 2017, 15(6):e2001855.
- Wenz B, Nielsen S, Gassowski M, Santos-Hovener C, Cai W, Ross RS, Bock CT, Ratsch BA, Kucherer C, Bannert N et al: High variability of HIV and HCV seroprevalence and risk behaviours among people who inject drugs: results from a cross-sectional study using respondent-driven sampling in eight German cities (2011-14). BMC Public Health 2016, 16(1):927.
- Plotzki E, Keller M, Ivanusic D, Denner J: A new Western blot assay for the detection of porcine cytomegalovirus (PCMV). J Immunol Methods 2016, 437:37-42.
- Pinkenburg O, Meyer T, Bannert N, Norley S, Bolte K, Czudai-Matwich V, Herold S, Gessner A, Schnare M: The Human Antimicrobial Protein Bactericidal/Permeability-Increasing Protein (BPI) Inhibits the Infectivity of Influenza A Virus. PLoS One 2016, 11(6):e0156929.
- Nielsen S, Gassowski M, Wenz B, Bannert N, Bock CT, Kucherer C, Ross RS, Bremer V, Marcus U, Zimmermann R et al: Concordance between self-reported and measured HIV and hepatitis C virus infection status among people who inject drugs in Germany. Hepatol Med Policy 2016, 1:8.
- Kramer P, Lausch V, Volkwein A, Hanke K, Hohn O, Bannert N: The human endogenous retrovirus K(HML-2) has a broad envelope-mediated cellular tropism and is prone to inhibition at a post-entry, pre-integration step. Virology 2016, 487:121-128.
- Ivanusic D, Denner J, Bannert N: Correlative Forster Resonance Electron Transfer-Proximity Ligation Assay (FRET-PLA) Technique for Studying Interactions Involving Membrane Proteins. Curr Protoc Protein Sci 2016, 85:29 17 21-29 17 13.
- Hohn O, Mostafa S, Norley S, Bannert N: Development of an antigen-capture ELISA for the detection of the p27-CA protein of HERV-K(HML-2). J Virol Methods 2016, 234:186-192.
- Hanke K, Hohn O, Bannert N: HERV-K(HML-2), a seemingly silent subtenant - but still waters run deep. APMIS 2016, 124(1-2):67-87.
- Gabriel B, Fiebig U, Hohn O, Plesker R, Coulibaly C, Cichutek K, Muhlebach MD, Bannert N, Kurth R, Norley S: Suppressing active replication of a live attenuated simian immunodeficiency virus vaccine does not abrogate protection from challenge. Virology 2016, 489:1-11.
- Semaan M, Ivanusic D, Denner J: Cytotoxic Effects during Knock Out of Multiple Porcine Endogenous Retrovirus (PERV) Sequences in the Pig Genome by Zinc Finger Nucleases (ZFN). PLoS One 2015, 10(4):e0122059.
- Schmier S, Mostafa A, Haarmann T, Bannert N, Ziebuhr J, Veljkovic V, Dietrich U, Pleschka S: In Silico Prediction and Experimental Confirmation of HA Residues Conferring Enhanced Human Receptor Specificity of H5N1 Influenza A Viruses. Sci Rep 2015, 5:11434.
- Rabenau HF, Bannert N, Berger A, Donoso Mantke O, Eberle J, Enders M, Fickenscher H, Grunert HP, Gurtler L, Heim A et al: [Not Available]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2015, 58(9):1025.
- Jurke A, Bannert N, Brehm K, Fingerle V, Kempf VA, Kompf D, Lunemann M, Mayer-Scholl A, Niedrig M, Nockler K et al: Serological survey of Bartonella spp., Borrelia burgdorferi, Brucella spp., Coxiella burnetii, Francisella tularensis, Leptospira spp., Echinococcus, Hanta-, TBE- and XMR-virus infection in employees of two forestry enterprises in North Rhine-Westphalia, Germany, 2011-2013. Int J Med Microbiol 2015, 305(7):652-662.
- Ivanusic D, Madela K, Laue M, Denner J: Three-Dimensional Imaging of CD63 Recruitment at the Virological Synapse: HIV-1. AIDS Res Hum Retroviruses 2015, 31(6):579-580.
- Ivanusic D, Heinisch JJ, Eschricht M, Laube U, Denner J: Improved split-ubiquitin screening technique to identify surface membrane protein-protein interactions. Biotechniques 2015, 59(2):63-73.
- Ivanusic D: In Sweet Harmony: CD63 and HIV-1. AIDS Res Hum Retroviruses 2015, 31(4):361-362.
- Hauser A, Kuecherer C, Kunz A, Dabrowski PW, Radonic A, Nitsche A, Theuring S, Bannert N, Sewangi J, Mbezi P et al: Comparison of 454 Ultra-Deep Sequencing and Allele-Specific Real-Time PCR with Regard to the Detection of Emerging Drug-Resistant Minor HIV-1 Variants after Antiretroviral Prophylaxis for Vertical Transmission. PLoS One 2015, 10(10):e0140809.
- Desimmie BA, Weydert C, Schrijvers R, Vets S, Demeulemeester J, Proost P, Paron I, De Rijck J, Mast J, Bannert N et al: HIV-1 IN/Pol recruits LEDGF/p75 into viral particles. Retrovirology 2015, 12:16.
- Zimmermann R, Marcus U, Schaffer D, Leicht A, Wenz B, Nielsen S, Santos-Hovener C, Ross RS, Stambouli O, Ratsch BA et al: A multicentre sero-behavioural survey for hepatitis B and C, HIV and HTLV among people who inject drugs in Germany using respondent driven sampling. BMC Public Health 2014, 14:845.
- Meixenberger K, Hauser A, Jansen K, Yousef KP, Fiedler S, von Kleist M, Norley S, Somogyi S, Hamouda O, Bannert N et al: Assessment of ambiguous base calls in HIV-1 pol population sequences as a biomarker for identification of recent infections in HIV-1 incidence studies. J Clin Microbiol 2014, 52(8):2977-2983.
- Madela K, Banhart S, Zimmermann A, Piesker J, Bannert N, Laue M: A simple procedure to analyze positions of interest in infectious cell cultures by correlative light and electron microscopy. Methods Cell Biol 2014, 124:93-110.
- Lausch V, Hermann P, Laue M, Bannert N: Silicon nitride grids are compatible with correlative negative staining electron microscopy and tip-enhanced Raman spectroscopy for use in the detection of micro-organisms. J Appl Microbiol 2014, 116(6):1521-1530.
- Ivanusic D, Madela K, Laue M, Denner J: Visualization of HIV-1 budding structures. AIDS Res Hum Retroviruses 2014, 30(10):945-946.
- Ivanusic D, Eschricht M, Denner J: Investigation of membrane protein-protein interactions using correlative FRET-PLA. Biotechniques 2014, 57(4):188-191, 193-188.
- Ivanusic D: HIV-1 cell-to-cell spread: CD63-gp41 interaction at the virological synapse. AIDS Res Hum Retroviruses 2014, 30(9):844-845.
- Hohn O, Hanke K, Lausch V, Zimmermann A, Mostafa S, Bannert N: CMV-promoter driven codon-optimized expression alters the assembly type and morphology of a reconstituted HERV-K(HML-2). Viruses 2014, 6(11):4332-4345.
- Hauser A, Santos-Hoevener C, Meixenberger K, Zimmermann R, Somogyi S, Fiedler S, Hofmann A, Bartmeyer B, Jansen K, Hamouda O et al: Improved testing of recent HIV-1 infections with the BioRad avidity assay compared to the limiting antigen avidity assay and BED Capture enzyme immunoassay: evaluation using reference sample panels from the German Seroconverter Cohort. PLoS One 2014, 9(6):e98038.
- Blochmann R, Curths C, Coulibaly C, Cichutek K, Kurth R, Norley S, Bannert N, Fiebig U: A novel small animal model to study the replication of simian foamy virus in vivo. Virology 2014, 448:65-73.
- Hohn O, Hanke K, Bannert N: HERV-K(HML-2), the Best Preserved Family of HERVs: Endogenization, Expression, and Implications in Health and Disease. Front Oncol 2013, 3:246.
- Hanke K, Hohn O, Liedgens L, Fiddeke K, Wamara J, Kurth R, Bannert N: Staufen-1 interacts with the human endogenous retrovirus family HERV-K(HML-2) rec and gag proteins and increases virion production. J Virol 2013, 87(20):11019-11030.
- Hanke K, Chudak C, Kurth R, Bannert N: The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int J Cancer 2013, 132(3):556-567.
- Girstmair H, Saffert P, Rode S, Czech A, Holland G, Bannert N, Ignatova Z: Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Rep 2013, 3(1):148-159.
- Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J et al: LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 2013, 10:57.
- Chudak C, Beimforde N, George M, Zimmermann A, Lausch V, Hanke K, Bannert N: Identification of late assembly domains of the human endogenous retrovirus-K(HML-2). Retrovirology 2013, 10:140.
nach oben