Research projects
Increasing the significance and public health benefit of the Integrated Genomic Surveillance of HIV by means of automations and extensions
As part of a BMG-funded project, the IGS-HIV (formerly MolSurv-HIV) is being further developed in order to continuously assess the German HIV epidemic and to enable an evidence-based design of public health measures in the areas of prevention, diagnostics and therapy. For this purpose, various laboratory and bioinformatic automations and extensions are planned by the end of 2025 (see Figure 1, marked in green). The project includes the full molecular recording of all new HIV diagnoses, for which the transmission of existing HIV sequences from the genotypic drug resistance testing in diagnostic laboratories together with the notification via DEMIS is being examined. The laboratory process in the HIV study laboratory is to be automated and sequencing extended to the full HIV genome. Automated bioinformatic pipelines are to be set up for the evaluation of the NGS data and the subsequent analyzes with the HIV sequences. This also includes the evaluation of a bioinformatic method for determining the recency or duration of infection in the InzSurv-HIV via the HIV-NGS sequence. Quarterly, the results on primary drug resistance, diversity, transmission clusters and outbreaks as well as the recency are to be automatically evaluated and presented to the public via a dashboard.

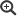 Figure 1: Planned workflow of the IGS-HIV. Corresponding extensions are marked in green. Source: © RKI
Figure 1: Planned workflow of the IGS-HIV. Corresponding extensions are marked in green. Source: © RKI
Evaluation of an automated AmpliSeq method for HIV sequencing as new HIV diagnoses increase due to the Ukraine war
As part of the IGS-HIV, around 60% of all new HIV diagnoses are currently being examined in the HIV study laboratory using molecular-biological techniques. The current laboratory process is based on the amplification and sequencing (NGS) of single HIV genome sections starting from viral RNA. However, the laboratory process is only partially automated and the labor- and time-consuming processing of the samples allows neither an increase in sample throughput nor an extension to the full HIV genome. The aim is therefore to automate the laboratory process for IGS-HIV, which will enable complete recording of the approximately 3,000-3,500 new HIV diagnoses in Germany every year. Against the background of the Ukrainian refugee movement, this goal is becoming even more important in order to be able to monitor the effects of the Ukraine war on the HIV epidemic in Germany as comprehensively as possible and to be able to initiate targeted prevention and public health measures.
The aim of the project is to set up an automated laboratory process that, with increasing sample volumes and consistently good sequence results, leads to a reduction in workload and thus an increase in cost-effectiveness. For this purpose, two methods for amplicon sequencing (AmpliSeq) from Thermo Fisher Scientific and Illumina are compared. Furthermore, an approach of viral capturing with Illumina sequencing is evaluated.
Detection of causes for the increase in HIV infections among injecting drug users in Germany
In Germany, outbreaks or rapidly growing sub-epidemics occur again and again, which are spatially limited but cannot be seen in general surveillance due to their relatively small number of cases. These clusters and sub-epidemics are often only noticed in later detailed analyses, which means that possible preventive measures are delayed or no longer applicable. These include, for example, clusters of people who inject drugs (PWID) in which the virus spreads locally over a short period of time. The early, automated detection of these fast-growing infection clusters would be enormously helpful in order to be able to take preventive action in the relevant regions and interrupt transmission paths at an early stage.
The aim of the project is to quickly detect new HIV sequences of fast-growing clusters in the respective transmission groups and to assign them accordingly. Due to the large amount of data, detection is automated based on the HIV-TRACE software. After automatic pre-sorting according to the subtypes of the sequences to be examined, these should be transferred to HIV-TRACE. An automated cluster allocation pipeline is to be established in which the various sub-epidemics of HIV are stored as reference data sets. Test data sets should be used to verify the correct assignment of sequences. Subsequently, sequences from 2021 and 2022 will be examined for membership of known PWID clusters in order to examine the extent to which outbreak events are responsible for the increase in reports in this risk group.
Development of a phylodynamic method for the analysis of HIV recombinants and characterization of HIV recombination events in the German epidemic
Recombination in multi- or superinfection is an essential feature of HIV and has an important impact on the evolution of the HIV epidemic. Individual URFs (unique recombinant forms) can result in epidemic-relevant CRFs (circulating recombinant forms) through successful retransmission. Further recombination of CRF creates complex recombinant forms. Molecular clock phylogenetic inference methods do not allow analysis of transmission networks and reproductive rates with recombinant HIV. In a doctoral project with the "Center for Artificial Intelligence in Public Health Research" (ZKI-PH 2, Dr. Denise Kühnert), a Bayesian method is to be set up that will make it possible to evaluate the transmission potential and the evolution of HIV recombinants.
Impact of HIV-related damage to the immune system on HIV/SARS-CoV-2 co-infection and SARS-CoV-2 vaccination (HIVCOV)
Compared to the general population, people with HIV show more severe COVID-19 courses and significantly lower concentrations of anti-SARS-CoV-2 IgG and neutralizing antibodies. The HIVCOV project therefore examined the vulnerability of people with HIV to SARS-CoV-2 infections and the effectiveness of anti-SARS-CoV-2 vaccines in order to contribute to the development of, for example, vaccination strategies for people with HIV.
First, the SARS-CoV-2 anti-S and anti-N positive cases were identified in the HIV-1 Seroconverter cohort in the period Q1/2020-Q3/2021. The relevant study participants were then invited to take part in the HIVCOV project in Q4/2021. Based on blood samples collected quarterly in 2022, the course of the humoral (anti-S IgG, neutralizing antibodies) and cellular immune responses (T cell activation) after a SARS-CoV-2 infection in the HIVCOV participants are currently being examined. At the same time, a clinical-epidemiological survey on SARS-CoV-2 vaccinations and infections was carried out from Q4/2021. In total, samples and data for the HIVCOV project were obtained from 251 and 1042 participants in the HIV-1 Seroconverter Study.
to the top