Grif K, Karch H, Schneider C, Daschner F D, Beutin L, Cheasty T, Smith H, Rowe B, Dierich M P, Allerberger F (1998): Comparative study of five different techniques for epidemiological typing of Escherichia coli O157
Diagn. Microbiol. Infect. Dis. 32: 165-176.
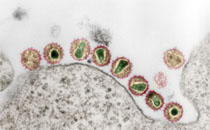
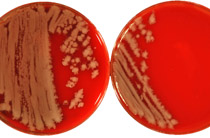
A set of 47 Austrian human, food, and veterinary Escherichia coli O157:H7 isolates was used to evaluate five different epidemiological typing methods. Ribotyping using an automated microbial characterization system (RiboPrinter) was not suitable for detection of epidemiological relatedness. All but one E. coli strain were typeable by phage typing. Random amplified polymorphic DNA-PCR fingerprinting was performed using primer M13 containing the sequence 5'-GAG GGT GGC GGT TCT-3' and primer 1247 (5'-AAGAGCCCGT-3'). Although both methods recognized only two clusters, both dendrograms grouped most of the EHEC O157 isolates into epidemiologically related subgroups. Pulsed-field gel electrophoresis of XbaI digested total DNA was a valuable subtyping system. We found that major differences can exist between results of multiple subtyping methods. E. coli O157 isolates should not be classified as epidemiologically related or nonrelated on the basis of a single typing method alone.