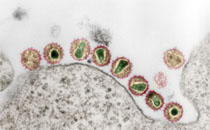
Ehlers B, Dural G, Yasmum N, Lembo T, de Thoisy B, Ryser-Degiorgis MP, Ulrich RG, McGeoch DJ (2008): Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: Cospeciation and interspecies transfer
J. Virol. 82 (7): 3509-3516.
Novel members of the subfamily Gammaherpesvirinae, hosted by eight mammalian species from six orders (Primates, Artiodactyla, Perissodactyla, Carnivora, Scandentia, Eulipotyphla), were discovered using PCR with pan-herpes DNA polymerase (DPOL) gene primers and genus-specific glycoprotein B (gB) gene primers. The gB and DPOL sequences of each virus species were connected by long-distance PCR and contiguous sequences of approximately 3.4 kbp compiled. Six additional gammaherpesviruses from four mammalian host orders (Artiodactyla, Perissodactyla, Primates, Proboscidea), for which only short DPOL sequences were known, were analysed in the same manner. Together with available corresponding sequences for 31 other gammaherpesviruses, alignments of encoded amino acid sequences were made and used for phylogenetic analyses by maximum-likelihood and Bayesian Monte Carlo Markov chain methods, to derive a tree which contained two major loci of unresolved branching detail. The tree was rooted by parallel analyses that included alpha- and betaherpesvirus sequences. This gammaherpesvirus tree contains eleven major lineages, and presents the widest view to date of phylogenetic relationships in any subfamily of the Herpesviridae, as well as the most complex in number of deep lineages. The tree’s branching pattern can only in part be interpreted in terms of cospeciation of virus and host lineages, and a substantial incidence of interspecies transfer of viruses must also be invoked.