Laverde Gomez JA, van Schaik W, Freitas AR, Coque TM, Weaver KE, Francia MV, Witte W, Werner G (2010): A multiresistance megaplasmid pLG1 bearing a hylEfm genomic island in hospital Enterococcus faecium isolates
Int. J. Med. Microbiol.: Epub Oct 15. doi:10.1016/j.ijmm.2010.08.015.
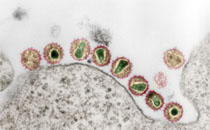
Enterococcus faecium is considered to be a nosocomial pathogen with increasing medical importance. The putative virulence factor, hylEfm, encoding a putative hyaluronidase, is enriched among the hospital-associated polyclonal subpopulation of E. faecium.. The hylEfm gene is described to be part of a genomic island and was recently identified to be plasmid-located. Here, we present a description of the structure, localization, and distribution of the putative pathogenicity factor hylEfm and its putative island among 39 clinical isolates and elucidate the composition and host range of pLG1, a hylEfm multiresistance plasmid of approximately 281.02 kb.
The hylEfm gene was located within a 17,824-bp element highly similar to the putative genomic island (GI) structure that had been previously described. This genomic region was conserved among 39 hylEfm-positive strains with variation in a specific region downstream of hylEfm in 18 strains. The putative hylEfm was located on large plasmids (150–350 kb) in 37 strains. pLG1 could be horizontally transferred into four different E. faecium recipient strains (n = 4) but not into E. faecalis (n = 3). Sequencing of pLG1 resolved putative plasmid replication, conjugation, and maintenance determinants as well as a pilin gene cluster, carbon uptake and utilization genes, heavy metal and antibiotic resistance clusters.
The hylEfm transferable plasmid pLG1 bears additional putative pathogenicity factors and antibiotic resistance genes. These findings suggest horizontal gene transfer of virulence factors and antibiotic resistance gene clusters by a single genetic event (conjugative transfer) which might be triggered by heavy antibiotic use common in health care units where E. faecium is increasingly prevalent.